Research
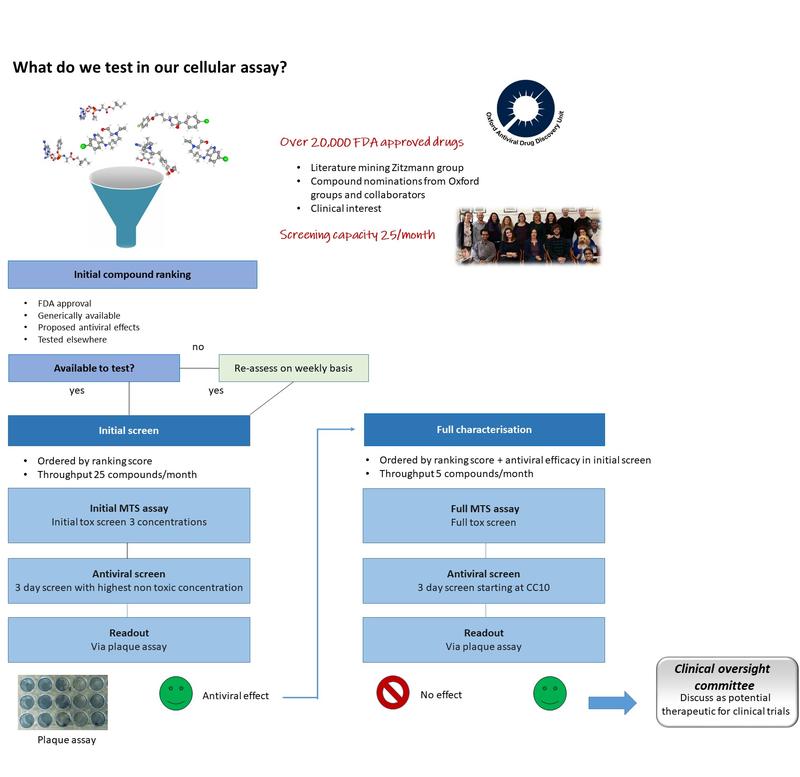
We are working with other groups across the globe to apply chemical principles and techniques to address the important biological questions around COVID-19. We are currently doing antiviral screens on hundreds of drugs and further details can be seen on our SARS CoV-2 Cellular Tracker.
For more details see the article about research in the Zitzmann lab during the pandemic and the 2022 Coronavirus Research Fund report.
Click here to learn more about host-targeting oral antiviral drugs to prevent pandemics.
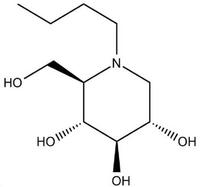
Iminosugars target host processes that many enveloped viruses depend on, hence they have the potential to be broad spectrum antivirals. Antiviral activity of iminosugar compounds has been demonstrated against viruses from 8 different virus families, including specifically dengue, hepatitis C virus, Ebola, influenza, HIV, hepatitis B viruses and others. We study the specific mechanisms of action of iminosugars against many of these viruses. We utilise virus-specific tools, reagents and models available to compare and tease out the specific proteins and pathways mediating the observed antiviral activity.
As iminosugars are potent inhibitors of alpha-glucosidase enzymes in the endoplasmic reticulum, we study their effects on viral (and host) protein misfolding and glycosylation. Certain iminosugars also inhibit glucocerebrosidase (their mechanism of action as a substrate reduction therapy in Gaucher and Niemann-Pick diseases), so in collaboration with Atsushi Kato (Japan) we test their broader enzyme inhibition profiles and try to form an integrated understanding of their effects on other pathways, such as lipogenesis.
What effects do iminosugars have on cells, both uninfected and in the context of pathogen infection? We use a primary human macrophage cell model of dengue virus infection, a hepatitis C virus cell culture system, in addition to HIV and influenza virus infection systems to investigate the key host and pathogen proteins involved in the anti-viral mechanism of action of iminosugars.
The calnexin cycle sub-group is a mixture of structural and molecular biologists looking at different aspects of the calnexin/calreticulin cycle, a subset of endoplasmic reticulum quality control (ERQC). This is a process by which the folding of glycoproteins is ordered and assessed by a series of lectins (glycan binding proteins). During or immediately following translation a large glycan can be added to certain asparagine residues in a process termed N-linked glycosylation. This glycan consists of two N-acetyl-glucosamines, nine mannoses and three terminal glucose residues. The three glucoses are trimmed down to just one by two enzymes termed α-glucosidase I & II. This monoglucosylated glycan is the substrate for calnexin and calreticulin, two proteins which complex with other chaperones such as ERp57 to aid in the folding of the monoglucosylated protein. α -Glucosidase II eventually removes this final glucose residue permitting release from calnexin. If the protein is correctly folded it proceeds to the Golgi for further processing, but if it is incorrectly folded then it can be re-glucosylated by an enzyme called UDP-glucose glycosyltransferase (UGGT 1), allowing it to re-bind calnexin/calreticulin and have another chance at folding correctly. Failure to fold correctly after a certain period leads to degradation of the glycoprotein by ER associated degradataion (ERAD).
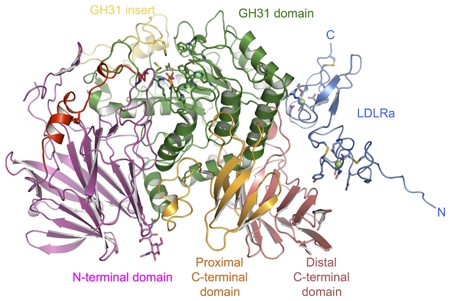
Crystal structure of mouse α-glucosidase II in cartoon representation
Eukaryotes have evolved in such a way that proteins normally destined for secretion are retained inside the cell if they are not properly folded. At the heart of this eukaryotic folding quality control system is the UDP-Glucose Glycoprotein Glucosyl Transferase aka UGGT, the single checkpoint enzyme in charge of flagging misfolded glycoproteins for retention.
Several mysteries surround UGGT. Starting from a gaze at its sequence, more than 1200 residues worth of completely unannotated sequence at the N terminus, make it nearly impossible to glean structural and functional understanding from bioinformatics alone.
We know that UGGT targets misfolded glycopeptides to chaperones inside the cell, by sticking a glucose moiety onto them, thus preventing them from prematurely entering their mature protein life.
But quite how this one protein can recognise misfolding of hundreds of different glycoproteins, and only glucosylate the ones that are not properly folded, is difficult to imagine. Nor is it clear if UGGT is only a checkpoint enzyme, or can instead also work with other cell chaperones to improve the folding of its unfortunate substrate glycoproteins.
And what if UGGT and its strict quality control were to do more harm than good to individuals carrying genetic mutations that only partially impair function? In some circumstances we can imagine it may be better for a partially active, slightly misfolded glycoprotein to reach its tissue destination, rather than remain trapped inside the cell by UGGT glucosylation.
We have started working towards the understanding of UGGT structure and function, and hunting for UGGT inihibitors of therapeutic potential. Watch this space!
We are studying the largely unknown structure, function, and location of the HCV p7 ion channel, a key target for drug development. We also study the structure of enzymes involved in protein folding, which shall allow us to further understand at the amino acid level the interactions between iminosugars and target proteins.
Proteomics involves the separation and identification of proteins in samples. We use proteomics to quantitatively compare protein expression among multiple samples, detect / quantify proteins in samples and identify proteins in samples / gel bands. The techniques we use in proteomics are two-dimensional gel electrophoresis (2D-PAGE) and mass spectrometry.
Currently we are mainly focusing on using proteomics to help discover novel liver fibrosis biomarkers. Liver biopsy is the reference standard for assessing liver fibrosis with several well-known disadvantages and no reliable non-invasive diagnostic approach is available to discriminate between the intermediate stages of fibrosis. We are using proteomics to identify (and measure levels of) novel fibrosis biomarkers in serum / plasma samples from patients with different degrees of liver fibrosis. We are analysing samples from patients with various liver diseases including hepatitis C. The biomarkers we have so far discovered look promising in comparison to the fibrosis biomarkers currently used in the clinic.
We are currently using mass spectrometry to identify more liver disease biomarkers using isotopically labelled tags which allows quantitative comparison of protein levels in multiple patient samples. We are also using mass spectrometry to detect and quantify these biomarkers in serum / plasma. Click here for more details on how the Zitzmann lab have used proteomics to identify potential novel protein biomarkers for fatty liver disease.
We have also used proteomics to investigate several viral systems including the identification of protein expression changes in cells infected with HCV, HBV, herpes simplex virus and also to investigate cell signalling in HIV.
Iminosugars are medicinally interesting compounds not only because of their broad spectrum antiviral ability, but also due to their capacity to influence a number of different cellular pathways and functions: protein folding, glycoprotein synthesis, glycolipid metabolism and others. While this variety of activities can be an asset, it is important to be able to design more specific iminosugars to minimize side effects without loss of desired activity.
The medicinal chemistry program currently focuses on a systematic study of iminosugars. We are carrying out hypothesis-driven science, with each compound that is synthesized and tested answering a specific question about structure-activity relationships. By understanding how their structure dictates biological activity, we can rationally design the next generation of antiviral compounds.
The strength of the program rests on its close integration with a number of the other subgroups in the lab. By both synthesizing and testing the compounds for HCV, dengue, HIV and BVDV antiviral activity in house, a quick feedback cycle enables efficient exploration of iminosugar space. Work with the structural team is pursuing the cocrystallization of lead iminosugars with their enzyme targets. With this atomistic understanding, strategic modifications can be made to the iminosugar to optimize specific aspects of the enzyme-ligand interaction.
In addition to our group, we collaborate with Prof. Atsushi Kato and his group (University of Toyama; Toyama, Japan) to evaluate the iminosugars’ ability to inhibit an array of isolated enzymes, while Dr. Dom Alonzi measures the iminosugars’ effects on glycoprotein processing and glycosphingolipid levels in cell culture. To complement the viruses we study, Prof. Norica Nichita-Branza (Institute of Biochemistry, Romanian Academy; Bucharest, Romania) investigates the potency of our test compounds against hepatitis B virus.
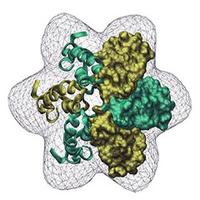
The exact role of p7 in the HCV life cycle is still unclear, but several studies indicate that p7 carries out more than one function. Recently published experiments from our laboratory have shown that the lipid environment affects structural features and the ion channel activity of p7. It is therefore possible that lipids play a role in modulating different functions of p7.
To understand how antiviral drugs acting on p7 disrupt the viral life cycle, it will be important to elucidate how lipids influence p7. In line with this concept we are using a set of biophysical approaches to further study how lipids affect p7 and its interaction with antiviral iminosugars.
We are trying to understand the significance of this process in the folding of gp120, the major surface glycoprotein of HIV, from an antiviral perspective. The specific antiviral effects of iminosugars on the folding of gp120 is being investigating as well as the potential of other components of the calnexin cycle, including UGGT, as antiviral targets.